Trans-acting siRNAs (ta-siRNAs) are a specific class of small RNA molecules that play a critical role in regulating gene expression in plants after the genes are transcribed into RNA. Although similar in function to small interfering RNAs (siRNAs), ta-siRNAs have a distinct production process that allows for more precise control over gene activity. This article explores the structure, function, and production mechanism of ta-siRNAs, highlighting their importance in plant development and physiology.
Structure and Similarities to siRNAs
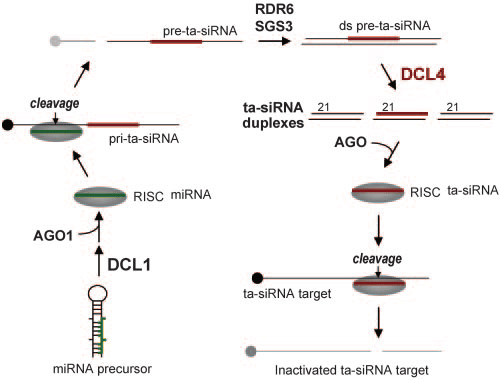
Like siRNAs, ta-siRNAs are short, double-stranded RNA molecules, typically 21 nucleotides long. They share a characteristic feature: 3' overhangs on both strands. These overhangs are essential for ta-siRNAs to integrate into a cellular complex called the RNA-induced silencing complex (RISC). This structural similarity allows ta-siRNAs to leverage the RNA interference (RNAi) machinery within the plant cell to cleave (cut) target messenger RNA (mRNA) molecules, ultimately silencing the genes they carry. Studying ta-siRNAs requires high-quality RNA extraction. Researchers can obtain reliable extraction tools from various suppliers.
Function: Precise Gene Silencing
Ta-siRNAs function by cleaving specific messenger RNA (mRNA) molecules in a sequence-dependent way. This differs from siRNAs, which originate from folded hairpin structures within the same RNA molecule. Instead, ta-siRNAs are produced from separate locations in the plant genome called trans-acting siRNA (TAS) genes [2]. This key difference allows ta-siRNAs to regulate genes anywhere in the genome, providing a more flexible and targeted approach to gene silencing. Researchers can utilize commercially available reagents From Maxanim for their studies on Ta-siRNA.
Production: A miRNA-Guided Process
The production of ta-siRNAs involves a multi-step process that relies on microRNAs (miRNAs) and specialized enzymes. First, precursor RNA molecules from TAS genes are cleaved by miRNAs in a process mediated by Argonaute proteins. This initial cleavage triggers the activity of RNA-dependent RNA polymerase 6 (RDR6), which converts the cleaved single-stranded RNA into double-stranded RNA (dsRNA). Finally, Dicer-like enzyme 4 (DCL4) processes the dsRNA into a series of mature ta-siRNAs, with each ta-siRNA having a slightly different sequence compared to its neighbors. This distinctive, "phased" nature of ta-siRNAs is a hallmark feature and a result of the stepwise processing by RDR6.
Conclusion
Trans-acting siRNAs are a powerful tool used by plants to fine-tune gene expression. Their unique production pathway, guided by miRNAs, allows for the precise regulation of non-overlapping mRNAs. Understanding the intricate mechanism of ta-siRNA action holds significant promise for unraveling plant development and uncovering novel strategies for improving crops.
Learn More by Watching This Video
Future Directions
Research on ta-siRNAs is ongoing, focusing on understanding the specific roles of different ta-siRNA families in various plant development processes. Additionally, exploring the potential for manipulating ta-siRNA production offers exciting possibilities for developing new tools in plant biotechnology.